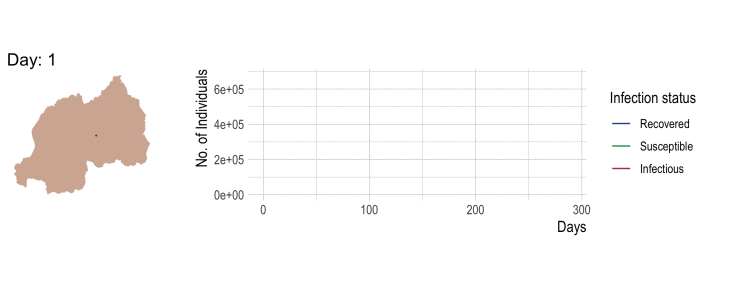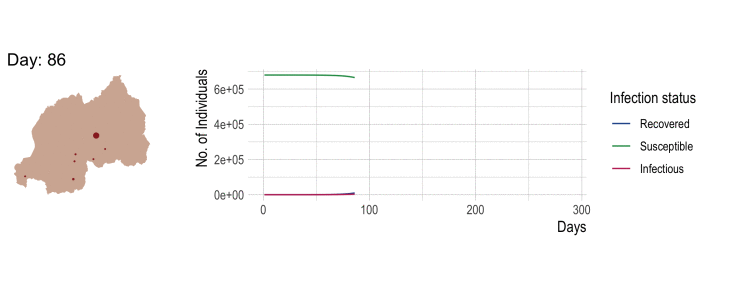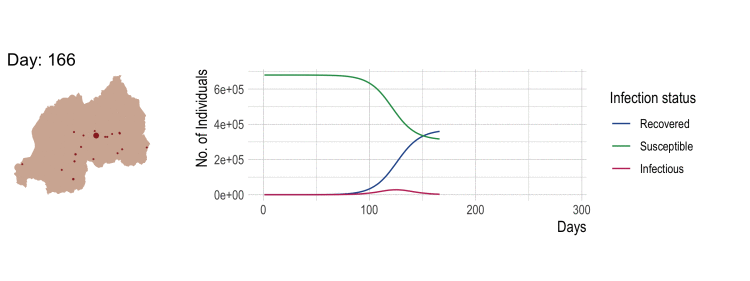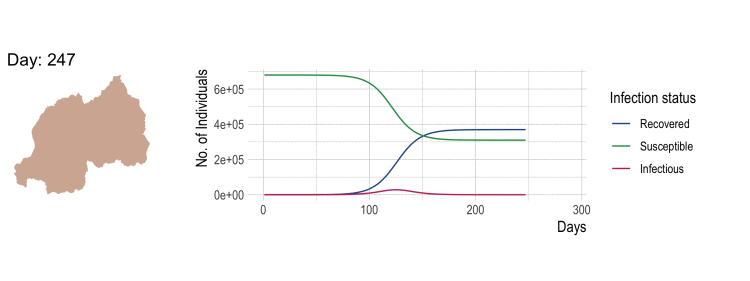
Compartmental disease model
Discrete time, stochastic, metatpopulation model.
In case of influenza, the compartmental model is SEIR:
S - Susceptible E - Latent I - Infectious R - Recovered
Main functions
disnet_commuting - Comuting function. Takes in network object (in graphml format), and calculates the commuting rates for all outgoing edges in network
disnet_sim_setup - Simulation setup function. Takes in the graph file with commuting rates added to it, and preps it up for runnning simulations.
disnet_simulate - Simulation function. This function runs the disease model simulations over the network.
library("disnet")
# Read in sample graph/network
f = system.file("sampleData", "g.rds", package = "disnet")
g = readRDS(f)
# calculate commuting rates over it
g_comm = disnet_commuting(g)
# select random node to seed infection in
set.seed(890)
nodes = igraph::vcount(g_comm)
seed_nd = igraph::vertex_attr(g_comm, "name", sample(1:nodes, 1))
# set up the network for simulations
for_sim = disnet_sim_setup(g_comm, seed_nd = seed_nd, output_dir = NA)
# run the simulations over the network
simres = disnet_simulate(sim_input = for_sim, sim_output_dir = NA)
simres2 = disnet_simulate(sim_input = for_sim, sim_output_dir = NA,
parallel = TRUE)